Software
Welcome to the ROCINATE (Really Original Collection of Interesting NANopore Tools for research Endevours)!
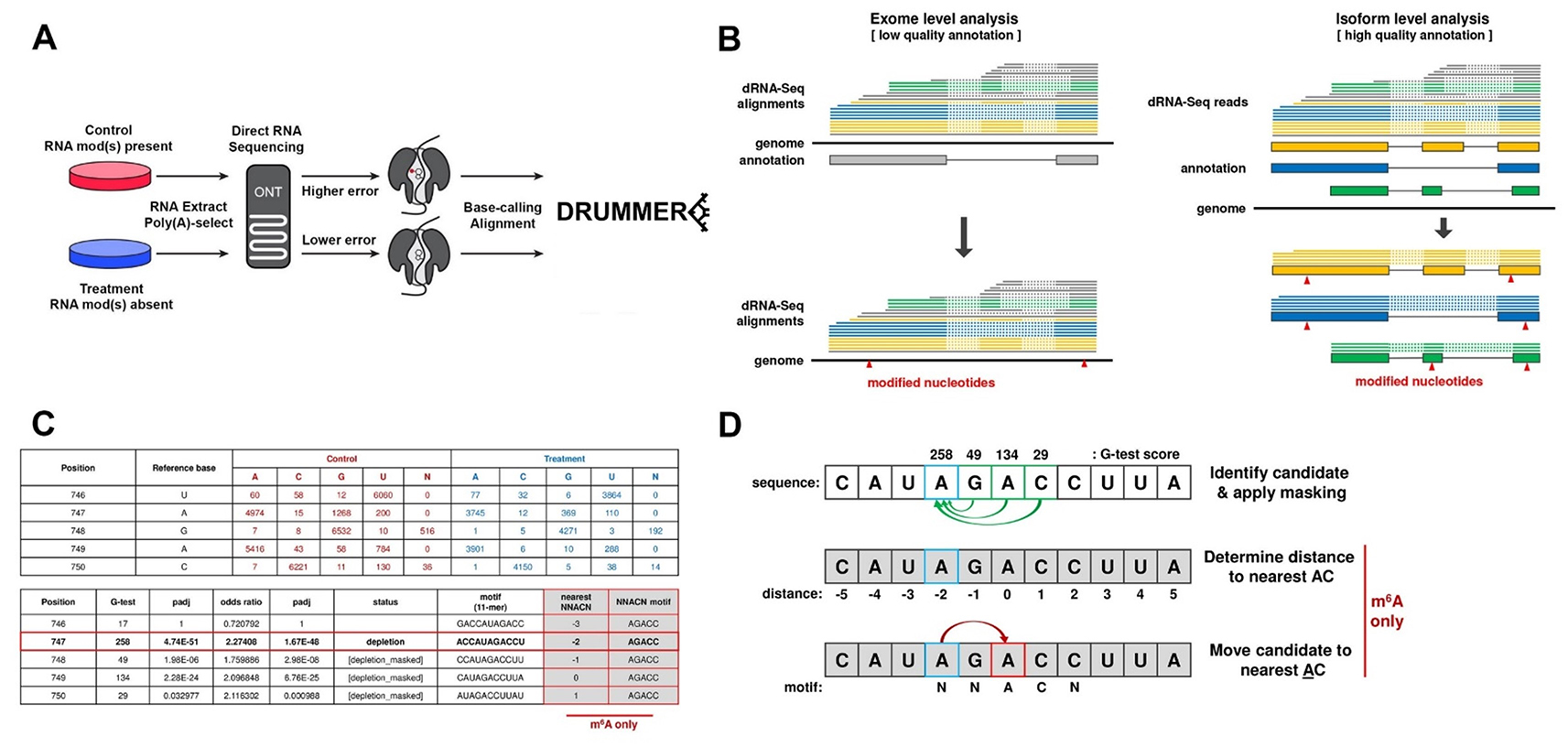
DRUMMER is designed to identify RNA modifications at nucleotide-level resolution on distinct transcript isoforms through the comparative analysis of basecall errors in Nanopore direct RNA sequencing (DRS) datasets.
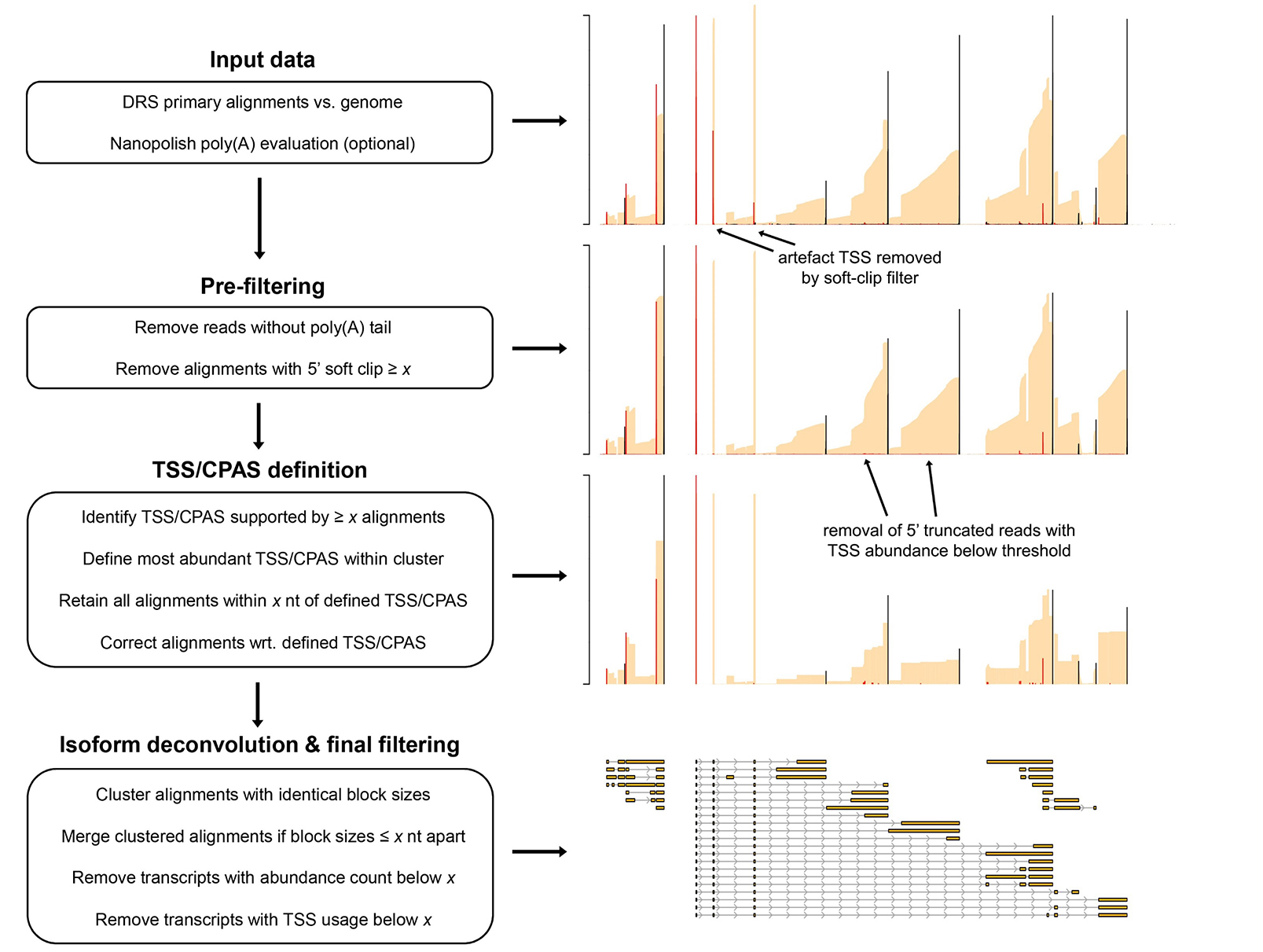
NAGATA uses Nanopore direct RNA sequencing reads aligned to a genome to produce a transcriptome annotation. NAGATA functions by parsing read alignments (sorted BAM files) to identify Transcription Units (TUs) by internally converting BAM into BED12 followed by numerically sorting ‘start’ and ‘end’ positions and then grouping alignments with similar ‘start’ and ‘end’ co-ordinates. This is performed on a row-by-row basis with a new TU defined only if the alignment co-ordinates of a given row differ from the previous row by greater than user-defined threshold (20 nt for transcription start sites (TSS), 50 nt for cleavage and polyadenylation sites (CPAS)).
