Publications
Highlighted
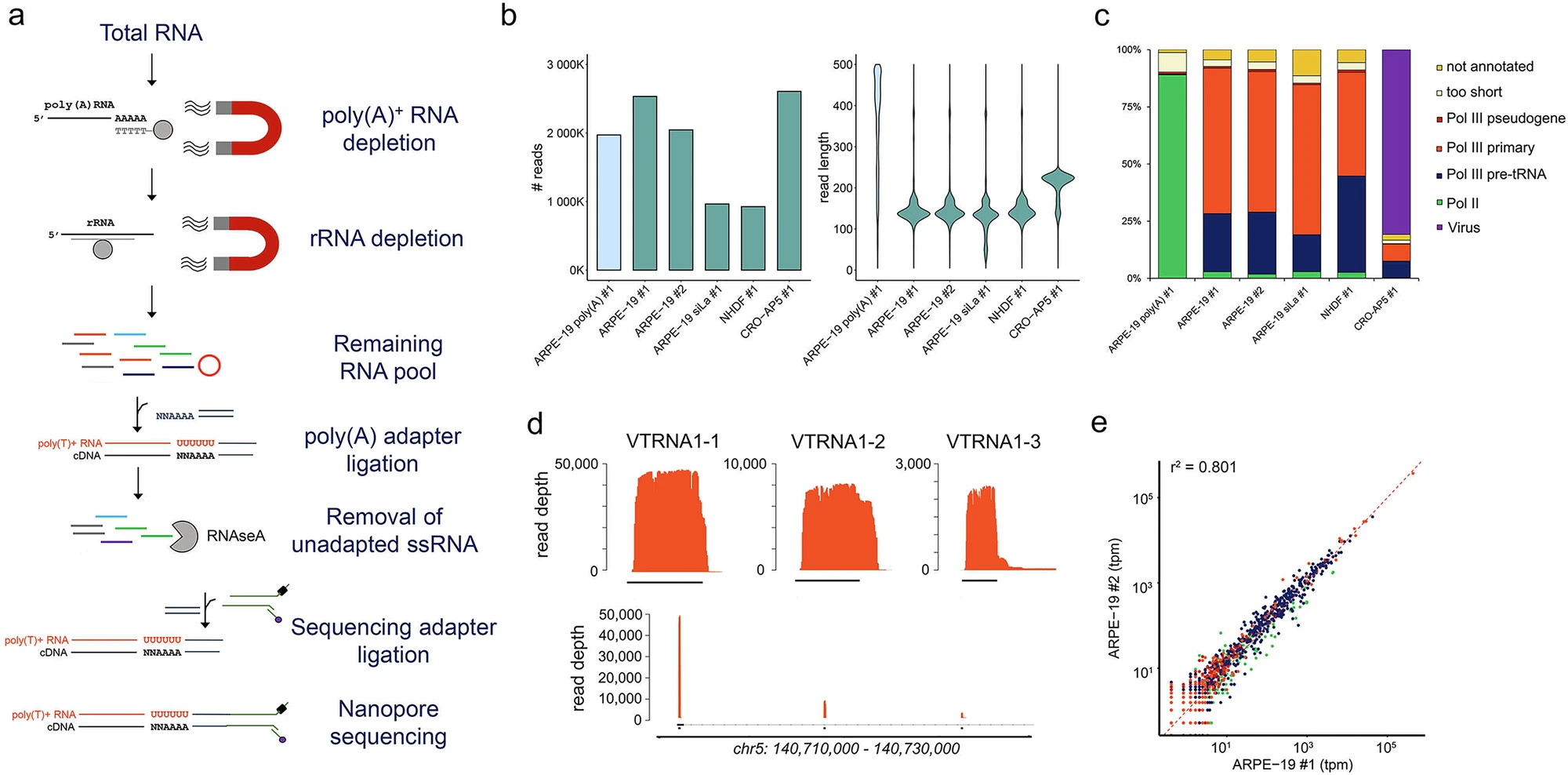
Defining expansions and perturbations to the RNA polymerase III transcriptome and epitranscriptome by modified direct RNA nanopore sequencing
Nature Communications
·
06 Jan 2026
·
doi:10.1038/s41467-025-68230-1
DRAP3R (Direct Read and Analysis of RNA Polymerase III derived RNAs) is a custom nanopore direct RNA sequencing method and analysis framework that enables the specific and sensitive capture of nascent Pol III-derived RNAs (and a small subset of Pol II-derived ncRNAs).
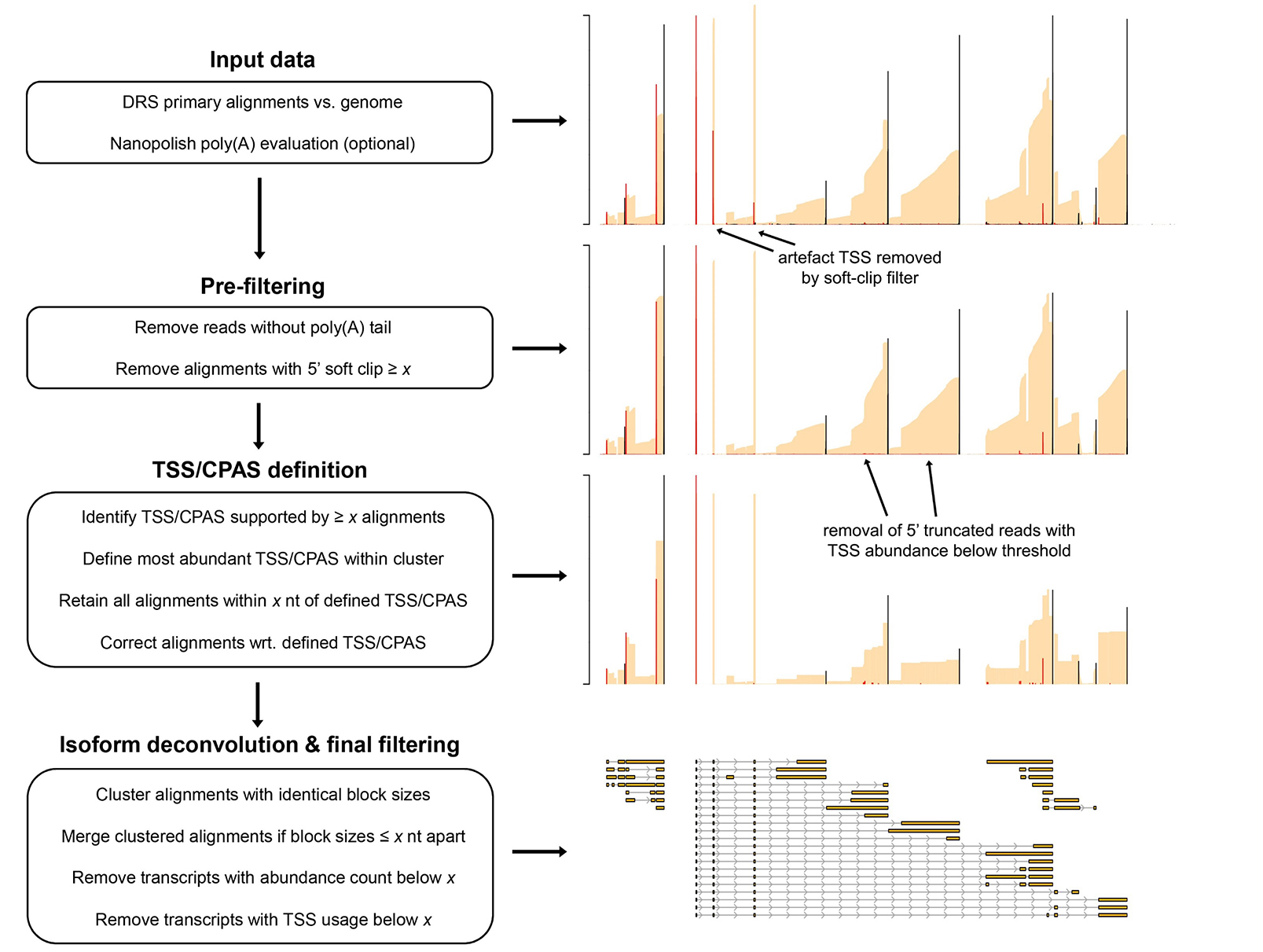
Nanopore guided annotation of transcriptome architectures
mSystems
·
02 Jul 2024
·
doi:10.1128/msystems.00505-24
NAGATA uses Nanopore direct RNA sequencing reads aligned to a genome to produce a transcriptome annotation. NAGATA functions by parsing read alignments (sorted BAM files) to identify Transcription Units (TUs) by internally converting BAM into BED12 followed by numerically sorting ‘start’ and ‘end’ positions and then grouping alignments with similar ‘start’ and ‘end’ co-ordinates. This is performed on a row-by-row basis with a new TU defined only if the alignment co-ordinates of a given row differ from the previous row by greater than user-defined threshold (20 nt for transcription start sites (TSS), 50 nt for cleavage and polyadenylation sites (CPAS)).
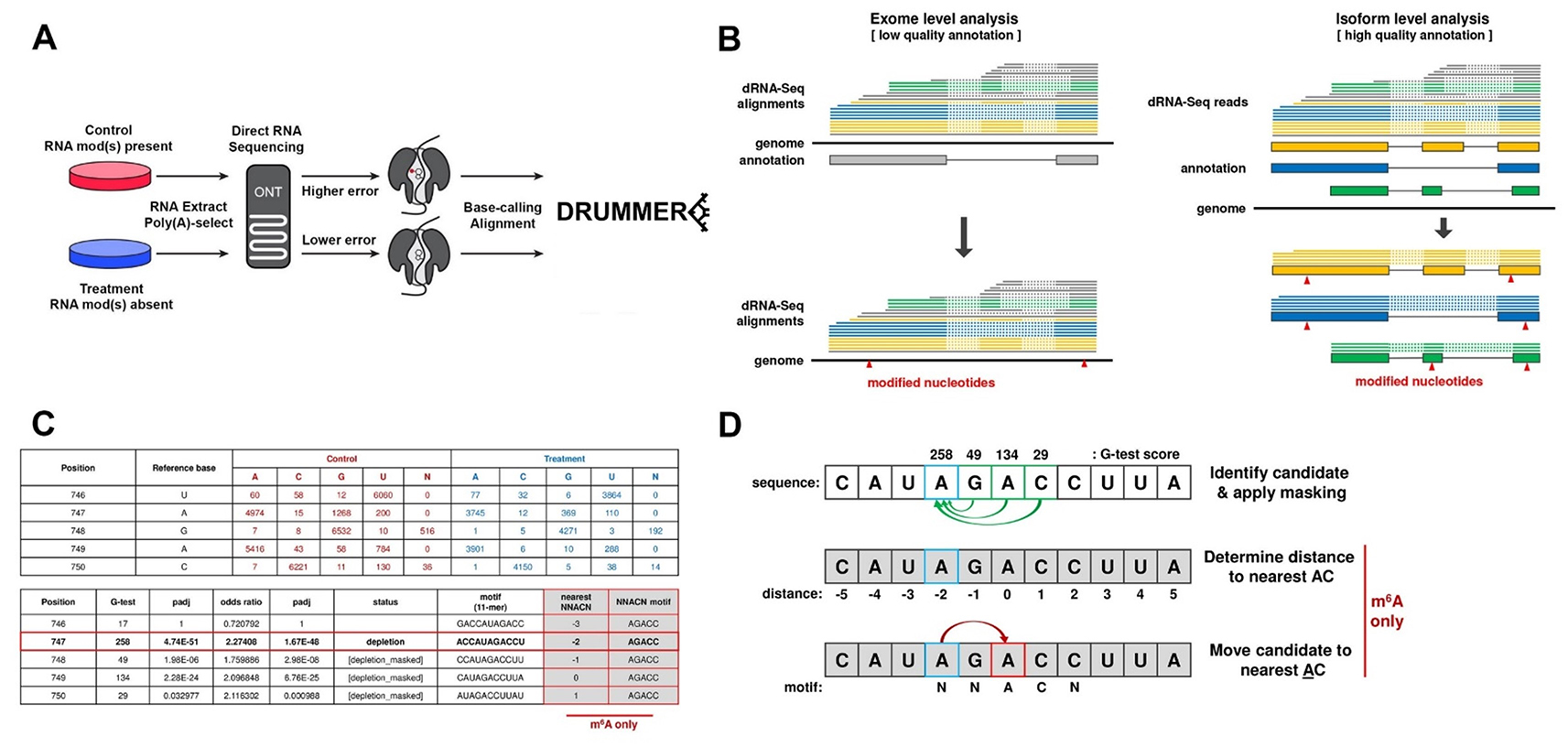
DRUMMER—rapid detection of RNA modifications through comparative nanopore sequencing
Bioinformatics
·
15 Apr 2022
·
doi:10.1093/bioinformatics/btac274
DRUMMER is designed to identify RNA modifications at nucleotide-level resolution on distinct transcript isoforms through the comparative analysis of basecall errors in Nanopore direct RNA sequencing (DRS) datasets.
All
2026

Defining expansions and perturbations to the RNA polymerase III transcriptome and epitranscriptome by modified direct RNA nanopore sequencing
Nature Communications
·
06 Jan 2026
·
doi:10.1038/s41467-025-68230-1
DRAP3R (Direct Read and Analysis of RNA Polymerase III derived RNAs) is a custom nanopore direct RNA sequencing method and analysis framework that enables the specific and sensitive capture of nascent Pol III-derived RNAs (and a small subset of Pol II-derived ncRNAs).
2025
Establishing benchmarks for quantitative mapping of m
6
A by Nanopore Direct RNA Sequencing
Cold Spring Harbor Laboratory
·
02 Nov 2025
·
doi:10.1101/2025.11.02.686099
RNA-interactome capture identifies SRSF3 as a key protein for herpesviral gene expression
PNAS Nexus
·
31 Jul 2025
·
doi:10.1093/pnasnexus/pgaf225
Detection of enterovirus RNA in pancreas and lymphoid tissues of organ donors with type 1 diabetes
Diabetologia
·
17 Mar 2025
·
doi:10.1007/s00125-025-06359-w
Defining expansions and perturbations to the RNA polymerase III transcriptome and epitranscriptome by modified direct RNA nanopore sequencing
Cold Spring Harbor Laboratory
·
12 Mar 2025
·
doi:10.1101/2025.03.07.641986
Sequence analysis of the hepatitis D virus across genotypes reveals highly conserved regions amidst evidence of recombination
Virus Evolution
·
01 Jan 2025
·
doi:10.1093/ve/veaf012
2024

Nanopore guided annotation of transcriptome architectures
mSystems
·
02 Jul 2024
·
doi:10.1128/msystems.00505-24
NAGATA uses Nanopore direct RNA sequencing reads aligned to a genome to produce a transcriptome annotation. NAGATA functions by parsing read alignments (sorted BAM files) to identify Transcription Units (TUs) by internally converting BAM into BED12 followed by numerically sorting ‘start’ and ‘end’ positions and then grouping alignments with similar ‘start’ and ‘end’ co-ordinates. This is performed on a row-by-row basis with a new TU defined only if the alignment co-ordinates of a given row differ from the previous row by greater than user-defined threshold (20 nt for transcription start sites (TSS), 50 nt for cleavage and polyadenylation sites (CPAS)).
Reply to Wang et al., “Ample evidence for the presence of HSV-1 LAT in non-neuronal ganglionic cells of mice and humans”
Journal of Virology
·
13 Jun 2024
·
doi:10.1128/jvi.00520-24
Reanalysis of single-cell RNA sequencing data does not support herpes simplex virus 1 latency in non-neuronal ganglionic cells in mice
Journal of Virology
·
16 Apr 2024
·
doi:10.1128/jvi.01858-23
Histone H2A variant H2A.B is enriched in transcriptionally active and replicating HSV-1 lytic chromatin
Journal of Virology
·
16 Apr 2024
·
doi:10.1128/jvi.02015-23
Nanopore Guided Annotation of Transcriptome Architectures
Cold Spring Harbor Laboratory
·
03 Apr 2024
·
doi:10.1101/2024.04.02.587744
tRNA epitranscriptome determines pathogenicity of the opportunistic pathogen
Pseudomonas aeruginosa
Proceedings of the National Academy of Sciences
·
07 Mar 2024
·
doi:10.1073/pnas.2312874121
2023
Histone H2A variant H2A.B is enriched in transcriptionally active HSV-1 lytic chromatin
Cold Spring Harbor Laboratory
·
22 Dec 2023
·
doi:10.1101/2023.12.22.573075
EMBO reports
·
17 Oct 2023
·
doi:10.15252/embr.202256327
Reanalysis of single-cell RNA sequencing data does not support herpes simplex virus 1 latency in non-neuronal ganglionic cells in mice
Cold Spring Harbor Laboratory
·
18 Jul 2023
·
doi:10.1101/2023.07.17.549345
2022
A Variant Allele in Varicella-Zoster Virus Glycoprotein B Selected during Production of the Varicella Vaccine Contributes to Its Attenuation
mBio
·
30 Aug 2022
·
doi:10.1128/mbio.01864-22
Nanopore-Based Detection of Viral RNA Modifications
mBio
·
28 Jun 2022
·
doi:10.1128/mbio.03702-21
DLK-Dependent Biphasic Reactivation of Herpes Simplex Virus Latency Established in the Absence of Antivirals
Journal of Virology
·
22 Jun 2022
·
doi:10.1128/jvi.00508-22
An eIF3d-dependent switch regulates HCMV replication by remodeling the infected cell translation landscape to mimic chronic ER stress
Cell Reports
·
01 May 2022
·
doi:10.1016/j.celrep.2022.110767

DRUMMER—rapid detection of RNA modifications through comparative nanopore sequencing
Bioinformatics
·
15 Apr 2022
·
doi:10.1093/bioinformatics/btac274
DRUMMER is designed to identify RNA modifications at nucleotide-level resolution on distinct transcript isoforms through the comparative analysis of basecall errors in Nanopore direct RNA sequencing (DRS) datasets.
2021
Single‐cell transcriptomics identifies Gadd45b as a regulator of herpesvirus‐reactivating neurons
EMBO reports
·
29 Nov 2021
·
doi:10.15252/embr.202153543
The architecture of the simian varicella virus transcriptome
PLOS Pathogens
·
22 Nov 2021
·
doi:10.1371/journal.ppat.1010084
Mutagenesis of the Varicella-Zoster Virus Genome Demonstrates That VLT and VLT-ORF63 Proteins Are Dispensable for Lytic Infection
Viruses
·
16 Nov 2021
·
doi:10.3390/v13112289
Widespread remodeling of the m
6
A RNA-modification landscape by a viral regulator of RNA processing and export
Proceedings of the National Academy of Sciences
·
19 Jul 2021
·
doi:10.1073/pnas.2104805118
Targeting the m6A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication
Genes & Development
·
24 Jun 2021
·
doi:10.1101/gad.348320.121
Enteric viruses evoke broad host immune responses resembling those elicited by the bacterial microbiome.
Cell host & microbe
·
23 Apr 2021
·
pmid:33894129
2020
Varicella-zoster virus VLT-ORF63 fusion transcript induces broad viral gene expression during reactivation from neuronal latency.
Nature communications
·
10 Dec 2020
·
pmid:33303747
The influence of human genetic variation on Epstein-Barr virus sequence diversity
Cold Spring Harbor Laboratory
·
04 Dec 2020
·
doi:10.1101/2020.12.02.20242370
Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing
Nature Communications
·
26 Nov 2020
·
doi:10.1038/s41467-020-19787-6
Decoding the Architecture of the Varicella-Zoster Virus Transcriptome
mBio
·
27 Oct 2020
·
doi:10.1128/mBio.01568-20
Expression of varicella-zoster virus VLT-ORF63 fusion transcript induces broad viral gene expression during reactivation from neuronal latency
Cold Spring Harbor Laboratory
·
11 Sep 2020
·
doi:10.1101/2020.09.11.294280
Analysis of the reiteration regions (R1 to R5) of varicella-zoster virus
Virology
·
01 Jul 2020
·
doi:10.1016/j.virol.2020.03.008
Using Direct RNA Nanopore Sequencing to Deconvolute Viral Transcriptomes.
Current protocols in microbiology
·
01 Jun 2020
·
pmid:32255550
Decoding the architecture of the varicella-zoster virus transcriptome
Cold Spring Harbor Laboratory
·
25 May 2020
·
doi:10.1101/2020.05.25.110965
Kallikrein-Mediated Cytokeratin 10 Degradation Is Required for Varicella Zoster Virus Propagation in Skin
Journal of Investigative Dermatology
·
01 Apr 2020
·
doi:10.1016/j.jid.2019.08.448
Evasion of a Human Cytomegalovirus Entry Inhibitor with Potent Cysteine Reactivity Is Concomitant with the Utilization of a Heparan Sulfate Proteoglycan-Independent Route of Entry.
Journal of virology
·
17 Mar 2020
·
pmid:31941787
Whole genome sequencing of Herpes Simplex Virus 1 directly from human cerebrospinal fluid reveals selective constraints in neurotropic viruses
Virus Evolution
·
01 Jan 2020
·
doi:10.1093/ve/veaa012
2019
Novel splicing and open reading frames revealed by long-read direct RNA sequencing of adenovirus transcripts
Cold Spring Harbor Laboratory
·
13 Dec 2019
·
doi:10.1101/2019.12.13.876037
Whole genome sequencing of Herpes Simplex Virus 1 directly from human cerebrospinal fluid reveals selective constraints in neurotropic viruses
Cold Spring Harbor Laboratory
·
06 Dec 2019
·
doi:10.1101/866350
Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing
Cold Spring Harbor Laboratory
·
05 Dec 2019
·
doi:10.1101/865485
Genetic and phenotypic intrastrain variation in herpes simplex virus type 1 Glasgow strain 17 syn+-derived viruses
Journal of General Virology
·
01 Dec 2019
·
doi:10.1099/jgv.0.001343
Chromatin dynamics and the transcriptional competence of HSV-1 genomes during lytic infections
PLOS Pathogens
·
14 Nov 2019
·
doi:10.1371/journal.ppat.1008076
Coevolution of Sites under Immune Selection Shapes Epstein-Barr Virus Population Structure.
Molecular biology and evolution
·
01 Nov 2019
·
pmid:31273385
Recurrent herpes zoster in the Shingles Prevention Study: Are second episodes caused by the same varicella-zoster virus strain?
Vaccine
·
01 Nov 2019
·
pmid:31679866
Human cytomegalovirus haplotype reconstruction reveals high diversity due to superinfection and evidence of within-host recombination.
Proceedings of the National Academy of Sciences of the United States of America
·
28 Feb 2019
·
pmid:30819890
Direct RNA sequencing on nanopore arrays redefines the transcriptional complexity of a viral pathogen
Nature Communications
·
14 Feb 2019
·
doi:10.1038/s41467-019-08734-9
Going the Distance: Optimizing RNA-Seq Strategies for Transcriptomic Analysis of Complex Viral Genomes
Journal of Virology
·
01 Jan 2019
·
doi:10.1128/JVI.01342-18
2018
Nuclear-cytoplasmic compartmentalization of the herpes simplex virus 1 infected cell transcriptome is co-ordinated by the viral endoribonuclease vhs and cofactors to facilitate the translation of late proteins
PLOS Pathogens
·
26 Nov 2018
·
doi:10.1371/journal.ppat.1007331
RNA m
Genes & development
·
21 Nov 2018
·
pmid:30463905
2018 Colorado Alphaherpesvirus Latency Society Symposium.
Journal of neurovirology
·
09 Nov 2018
·
pmid:30414047
High Viral Diversity and Mixed Infections in Cerebral Spinal Fluid From Cases of Varicella Zoster Virus Encephalitis.
The Journal of infectious diseases
·
05 Oct 2018
·
pmid:29986093
Use of Whole-Genome Sequencing of Adenovirus in Immunocompromised Pediatric Patients to Identify Nosocomial Transmission and Mixed-Genotype Infection.
The Journal of infectious diseases
·
08 Sep 2018
·
pmid:29917114
Native RNA sequencing on nanopore arrays redefines the transcriptional complexity of a viral pathogen
openRxiv
·
20 Jul 2018
·
doi:10.1101/373522
Molecular Aspects of Varicella-Zoster Virus Latency.
Viruses
·
28 Jun 2018
·
pmid:29958408
A spliced latency-associated VZV transcript maps antisense to the viral transactivator gene 61
Nature Communications
·
21 Mar 2018
·
doi:10.1038/s41467-018-03569-2
Comparative genomic, transcriptomic, and proteomic reannotation of human herpesvirus 6.
BMC genomics
·
20 Mar 2018
·
pmid:29554870
2017
Acute Retinal Necrosis Caused by the Zoster Vaccine Virus.
Clinical infectious diseases : an official publication of the Infectious Diseases Society of America
·
29 Nov 2017
·
pmid:29020238
A unique spliced varicella-zoster virus latency transcript represses expression of the viral transactivator gene 61
openRxiv
·
10 Aug 2017
·
doi:10.1101/174797
2016
Deep Sequencing of Distinct Preparations of the Live Attenuated Varicella-Zoster Virus Vaccine Reveals a Conserved Core of Attenuating Single-Nucleotide Polymorphisms.
Journal of virology
·
12 Sep 2016
·
pmid:27440875
Detection of Low Frequency Multi-Drug Resistance and Novel Putative Maribavir Resistance in Immunocompromised Pediatric Patients with Cytomegalovirus.
Frontiers in microbiology
·
09 Sep 2016
·
pmid:27667983
Viral Genome Sequencing Proves Nosocomial Transmission of Fatal Varicella.
The Journal of infectious diseases
·
28 Aug 2016
·
pmid:27571904
Islands of linkage in an ocean of pervasive recombination reveals two-speed evolution of human cytomegalovirus genomes.
Virus evolution
·
15 Jun 2016
·
pmid:30288299
In vitro system using human neurons demonstrates that varicella-zoster vaccine virus is impaired for reactivation, but not latency.
Proceedings of the National Academy of Sciences of the United States of America
·
12 Apr 2016
·
pmid:27078099
2015
Recombination of Globally Circulating Varicella-Zoster Virus.
Journal of virology
·
01 Jul 2015
·
pmid:25926648
Rapid Whole-Genome Sequencing of Mycobacterium tuberculosis Isolates Directly from Clinical Samples.
Journal of clinical microbiology
·
13 May 2015
·
pmid:25972414
Complete Genome Sequence of the Human Herpesvirus 6A Strain AJ from Africa Resembles Strain GS from North America.
Genome announcements
·
12 Feb 2015
·
pmid:25676750
Rates of vaccine evolution show strong effects of latency: implications for varicella zoster virus epidemiology.
Molecular biology and evolution
·
06 Jan 2015
·
pmid:25568346
2014
Whole-genome enrichment and sequencing of Chlamydia trachomatis directly from clinical samples.
BMC infectious diseases
·
12 Nov 2014
·
pmid:25388670
Evolution of cocirculating varicella-zoster virus genotypes during a chickenpox outbreak in Guinea-Bissau.
Journal of virology
·
01 Oct 2014
·
pmid:25275123
2013
Deep sequencing of viral genomes provides insight into the evolution and pathogenesis of varicella zoster virus and its vaccine in humans.
Molecular biology and evolution
·
25 Oct 2013
·
pmid:24162921
Genome Sequence of Human Herpesvirus 7 Strain UCL-1.
Genome announcements
·
10 Oct 2013
·
pmid:24115547
Mode of virus rescue determines the acquisition of VHS mutations in VP22-negative herpes simplex virus 1.
Journal of virology
·
17 Jul 2013
·
pmid:23864617
Next-generation whole genome sequencing identifies the direction of norovirus transmission in linked patients.
Clinical infectious diseases : an official publication of the Infectious Diseases Society of America
·
03 May 2013
·
pmid:23645848
Viral population analysis and minority-variant detection using short read next-generation sequencing.
Philosophical transactions of the Royal Society of London. Series B, Biological sciences
·
04 Feb 2013
·
pmid:23382427
2012
Correction: Specific Capture and Whole-Genome Sequencing of Viruses from Clinical Samples
PLoS ONE
·
24 Jan 2012
·
pmc:PMC3267775
2011
Specific capture and whole-genome sequencing of viruses from clinical samples.
PloS one
·
18 Nov 2011
·
pmid:22125625
Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania.
Genome research
·
28 Oct 2011
·
pmid:22038252
2010
Leishmania-specific surface antigens show sub-genus sequence variation and immune recognition.
PLoS neglected tropical diseases
·
28 Sep 2010
·
pmid:20927190
2009
TriTrypDB: a functional genomic resource for the Trypanosomatidae.
Nucleic acids research
·
20 Oct 2009
·
pmid:19843604
Comparative expression profiling of Leishmania: modulation in gene expression between species and in different host genetic backgrounds.
PLoS neglected tropical diseases
·
07 Jul 2009
·
pmid:19582145
2007
Comparative genomic analysis of three Leishmania species that cause diverse human disease.
Nature genetics
·
17 Jun 2007
·
pmid:17572675
RepSeq--a database of amino acid repeats present in lower eukaryotic pathogens.
BMC bioinformatics
·
11 Apr 2007
·
pmid:17428323
2005
COPASAAR--a database for proteomic analysis of single amino acid repeats.
BMC bioinformatics
·
03 Aug 2005
·
pmid:16078990
